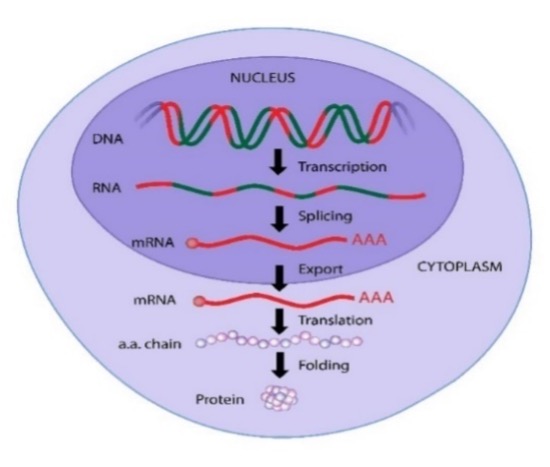
Our genome (DNA) codes for some 20 000 proteins. The DNA is tightly folded with its proteins (histones) to form nucleosomes that are snugly packaged chromatin. When a protein is needed the chromatin unfolds at exactly the right locations to let RNA in so it can translate the DNA to messenger RNA that carries the information outside the nucleus to the protein factory, the ribosome.
By Nils-Ola Holtze, Swedish retired surgent, stationed in Portugal
The mRNA consists of four different nucleotides, Adenine, Uracil, Cytosine, and Guanine ( AUCG) a combination of three (a codon) corresponding to a specific amino acid. Humans have 20 different amino acids that are the building blocks of every different protein. A human protein can consist of 44 to 27 000 amino acids.
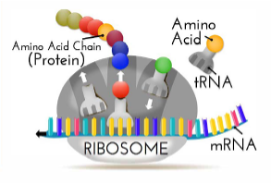
In the protein factory, the ribosomes, and the amino acid are assembled as a long chain and when done the amino acids fold to their 3-dimensional structure. This 3D structure is determining the protein’s function! Imagine you’re twisting a rubber band in opposite directions at its ends, when you move your hands together the rubber band folds into a 3D structure.
The problem arises that there are (almost) infinite alternatives for the protein to fold in space, e.g., in a protein with 100 amino acids the protein has 2 to the power of 100 alternative ways to fold, and only one of them gives the correct structure corresponding to its right function. And this happens in a second or less.
If we go back to the twisted rubber band. If you work fast, like putting your hands together each picosecond (a p-second is one trillionth of a second) for as long as the universe existed (allegedly 13,7 Billion years), the chances you get the same 3D structure twice are very minuscule.
If one calculates the alternatives for a protein with 1100 amino acids the ways it can fold are more than a centillion different ways, a centillion is 1 followed by 303 zeroes! And misfolding can lead to diseases as we see in mad cow disease.
Do you see the problem? How does the protein find its functional form in a second or less? And yet it happens billion times each day in our cells! This dilemma was formulated 50 years ago by Levinthal and remains a fascinating problem.
Read more by Holtze: Evolving Sience: Pseudomonas Syringae Bacteria Can Cause Lightning and Thunder
To put the problem in a recent context. Many of us have been injected with new mRNA or DNA vaccines. The idea is to make our own cells produce a modified spike protein. They had to modify it to make our cells accept it and to “stabilize” it so it wouldn’t break down easily.
To manage this, they had to tinker with the RNA nucleotides, the codons. This synthetic, modified protein consists of about 1400 amino acids that would represent 10 to the power of more than 600 (my calculator broke down crying) alternative ways to fold!
Since we have 20 different amino acids to build proteins and there are about 100 different combinations of every three combinations of the mRNA nucleotides, some codons translate to the same amino acids.
These are called synonymous codons. So, when we translate the codons in the ribosomes to amino acids, we can use different synonymous codons to get the same amino acid sequence that makes the final protein.
But the synonymous codons have different occupation times in the ribosomes which leads to different angles between the amino acids and thereby the 3D structure of the final protein. And structure is imperative to a protein’s function!
- So, my question is this; How on earth could anybody predict the 3D structure of this never-before-seen protein?
- And if by some miracle they could, who can say how it will react and interact in our bodies?
The theory is to teach our white blood cells to produce antibodies against this strange and never seen protein.
If our antibodies manage to fight down the protein, and hopefully no cross-reaction has taken place, it’s time for another booster. But of course, as usual, we must trust “The Science”.
Proteins are complicated structures. The structure in space determines its function. If something goes wrong it can cause inflammations, cancer, autoimmune diseases, thrombosis, accumulating in different tissues like the brain. And a dysfunctional protein doesn’t perform its designated function.
This paradox illustrates the immense complexity and enigma that is “life”.
- How could our proteins have evolved to the right shape and function to act and interact in an intricate way with other structures in our bodies?
Darwin told us that evolution progresses by random mutations. Mathematics tells us that a random evolution is impossible, there simply is not enough time even to make even one small functional, interacting protein.
Random probability theory tells us that if you give a monkey infinite time and a typewriter it would eventually be able to flawlessly reproduce Shakespeare’s complete works. I kind of doubt that. First, monkeys don’t live more than about 30 years. And secondly, I don’t believe in infinite, it doesn’t make any sense.
- Where does finite end and infinity begin? Is there a no-man’s-land between them?
- At what point does finite fall into the void of infinity?
Anyway, we seem to need (almost?) infinite time to randomly evolve even one small functional protein, and with a new random mutation, and another infinite time for it to evolve to something useful that can interact in a meaningful way.
And protein synthesis is even more complicated. We have sequenced about 2% of our protein-coding genes, much of the rest 98% are non-protein coding that we don’t really know the exact function of. Some decades ago, it was referred to as “junk DNA”.
The protein-coding DNA sequence consists mainly of two parts, Introns and Exons. The exons are what finally translate into a protein in the ribosomes. Intron sequences are interspersed nucleotides between the exons, like exon-intron-e-i-e-i-e-i-e-i-e.
When DNA transcribes to messenger-RNA, the introns are spliced out from the sequence and stay IN the nucleus. The EXons EXIT the nucleus in the form of mRNA to the ribosomes located in the cytoplasm, outside the nucleus.
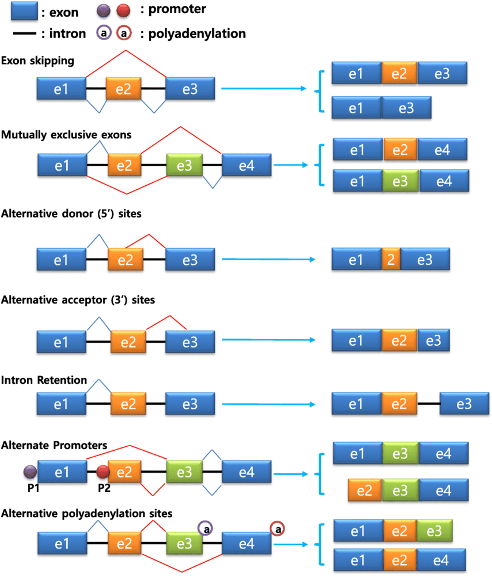
Now, when assembling the exons to a protein-coding mRNA sequence, introns are cut out. The cell has many different alternatives to put the exons together e.g., exon1-exon2-exon3-exon4-exon5, which can alternatively combine to exon1-exon3-exon2-exon-4-exon5.
This way we can produce similar but different proteins from the same gene. We can also exclude an exon or duplicate one. The possibilities are legion.
This process is called Gene-splicing, we know little about how the cell decides to splice a gene in a certain way. There are some fringe theories about why our cells behave like this, but my firm belief is that nothing in our cells or in our bodies is obsolete, or random, everything has a meaning and a function!
Many scientists seem to dismiss things they can’t explain as “junk” or a “fossil remains from an evolutionary past”.
So, the protein-making process is immensely complicated and we have much yet to discover. Many safety nets and control stations are in place, but if anything goes wrong the result could be a misfolded protein that can cause diseases such as prion disease In Creutzfeldt Jacobs disease and many neurological diseases.
If Darwin was wrong how then did, we evolve? I don’t know. Still to this day, it’s almost impossible, to make a career in academia if you don’t embrace The Darwinian Dogma. Imo Darwin described breeding in an unnecessarily complicated way, and extrapolated his findings way too far into evolution!
Epigenetics is an interesting subject. With it, the question of what we can do to evolve ourselves to meet our and our offspring’s optimal potential. Is it possible we all could live a long healthy life without medicines? We will discuss Epigenetics later.
By Nils-Ola Holtze, Swedish retired surgent, stationed in Portugal

